You're reading an old version of this documentation!
For the latest release, please have a look at v1.0.
3. How To Run#
3.1. Running G4SEE application#
Run the g4see application from command-line.
g4see [-h] [-v] /path/to/input/macro.mac [-o OUTPUT_FOLDER]
A macro file path is a mandatory input argument, see Input Macro files how to create macro files.
Output directory can be either relative or absolute path.
Examples:
Without any optional arguments:
g4see input.macDefine output directory:
g4see input.mac -o output/Redirect standard output (stdout) into a file:
g4see input.mac > log.txt
3.2. Merging histograms#
For single runs only (not for parametric runs), one can run the compiled C++ tool mergeHistograms.
This tool sums Edep and Ekin standard scoring histogram files in single folder or multiple folders (see below) into one histogram, as well as calculates standard deviation of each bin value.
mergeHistograms [-h] [-v] [/common/path/to/data/folder(_#)] [-o OUTPUT_FOLDER] [-d]
For the list of arguments and help, please run mergeHistograms -h.
Merging histogram files located in a single folder: define the path of that single folder
Merging histogram files located in multiple folders: define the common path ending with
#placeholder symbol (e.g.path/to/folder_#)If no folder path provided, by defualt the application will look for
out_#folders in current directory:out_0,out_1,out_2, etc.For details about output files and merging, see Output Files
For merging output files of parametric runs recursively, see Merging histograms recursively below
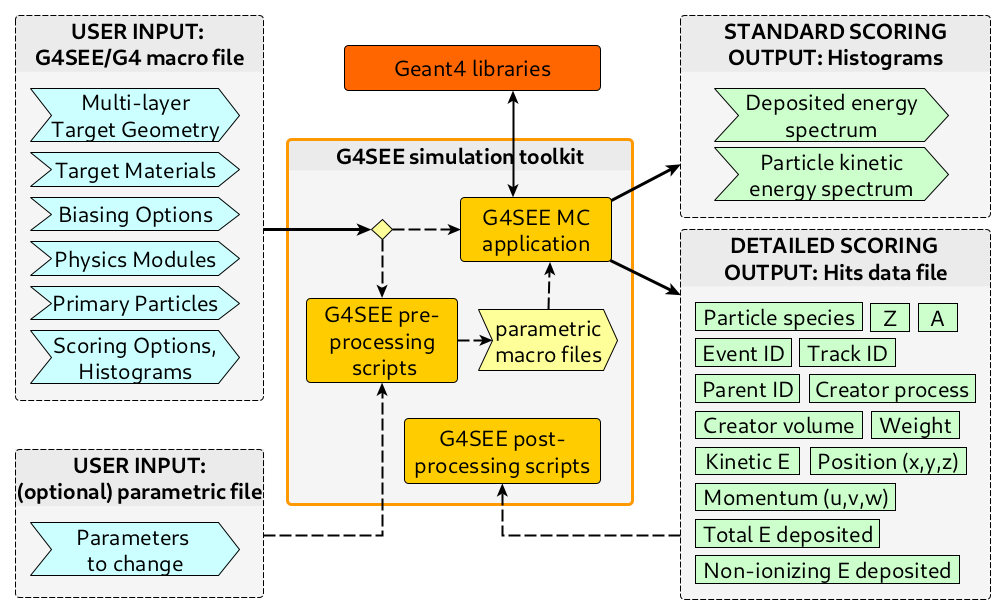
High-level architecture of the G4SEE toolkit#
3.3. Using G4SEE scripts#
In the build directory of G4SEE, there are useful auxiliary python3 scripts to start or delete jobs on a computer cluster, merge and visualize histogram data generated.
python3 g4see.py [-h] {submit,delete,view,merge,plot} ...
3.3.1. Submitting cluster jobs & parametrization#
Submitting G4SEE parallel jobs to a computer cluster queue for a single run using g4see.py submit.
Parallel jobs are jobs using the same macro file as input, but running on different computer cluster nodes independently
(with different random seeds if no seed defined in macro), to enhance performance and make use of a computer cluster.
Multi-threading and multi-processing are not necessarily supported on computer clusters,
therefore this script does not start multi-threading or multi-processing simulation runs.
python3 g4see.py submit [-h] -o OUTPUT -j JOBS [-q QUEUE] [-G4 G4_PATH] [-G4SEE G4SEE_PATH] input
For the list of arguments and help, please run python3 g4see.py submit -h.
The input can be either standard macro (.mac) or a parametric YAML (.yaml) files as well.
See Input Macro files how to create these macro files.
When using a parametric YAML (.yaml) file, a set of differently parametrized macro input files are automatically generated from a master macro file,
in order to perform parametric studies changing a single or even combination of multiple parameters/settings in the macro.
If
-q, --queueargument is provided, the script will submit the jobs to the defined cluster queue, otherwise it only generates the parametrized macros and output foldersThe number of parallel jobs defined via the
-j, --jobsargument (in parametric runs this is not the total job number, but the jobs per parameter values)Output folders defined with
-o, --outputare created automatically (in parametric runs folder names are based on the elements of thevalue-listkeyword)Already existing output folders (including all files) will be overwritten
If
g4seeis not installed inPATHenv. variable (e.g. in/bin/usr/local/), then G4SEE app’s absolute path should be provided via the-G4SEE, --G4SEE_PATHarg (default isg4see)If
geant4.shscript was not sourced (which is needed on cluster nodes), then Geant4 absolute path should be provided via the-G4, --G4_PATHarg (default isNone)
python3 g4see.py submit parametric_source.yaml -o outputs/ -j 10 -q normal
(This example submits 3*10=30 parametrized jobs in total to the cluster queue called “normal”.)
examples_parametric/parametric_source.yamlexamples_parametric/parametric_physics.yamlexamples_parametric/parametric_SV.yaml
3.3.2. Deleting cluster jobs#
To delete a set or all of a user’s G4SEE jobs submitted to computer cluster nodes. Only those jobs affected, which names start with “g4see”.
python3 g4see.py delete [-h] [-a] [-st {R,running,Q,queue}] [-q QUEUE] [-ss SUBSTRING] [-id left [right ...]] user
For the list of arguments and help, please run python3 g4see.py delete -h.
3.3.3. Visualizing geometry#
Providing a macro file as macro, you can visualize your geometry in 2D for checking or to include in documentation, presentation or paper.
python3 g4see.py view [-h] [-s SHOW] [-o OUTPUT] macro
For the list of arguments and help, please run python3 g4see.py view -h.
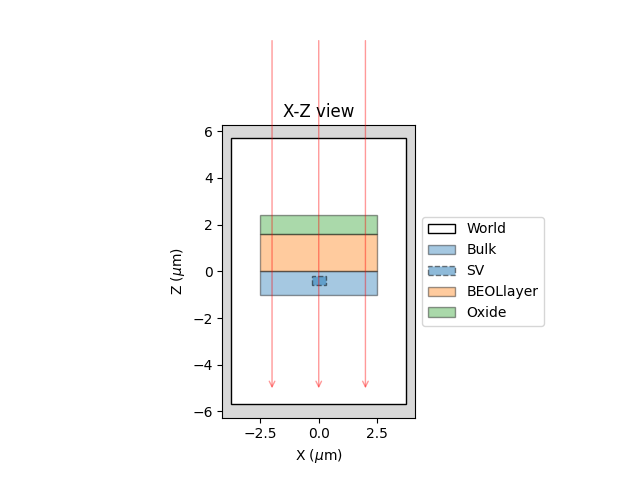
X-Z side view of target defined in SRAM example macro#
3.3.4. Merging histograms recursively#
After a parallel run, when you have many data files in separate folders, you need to merge histograms manually after all your jobs are finished. For parametric runs or multiple parallel jobs, you have recursive folder structure (set of parallel jobs), so you need to run:
python3 g4see.py merge [-h] folder
For the list of arguments and help, please run python3 g4see.py merge -h.
The script will merge all the data per run locally, so only folders with names out_# within each subdirectory.
This script calls mergeHistograms C++ tool (see Merging histograms above).
3.3.5. Plotting histograms#
Users can automatically generate a plot of one or multiple <hist>_<id>_histogram.out histogram files together.
python3 g4see.py plot [-h] [-ht HIST] [-o OUTPUT] [-nf NORMFACTOR] [-nb NORMBIN] data
For the list of arguments and help, please run python3 g4see.py plot -h.
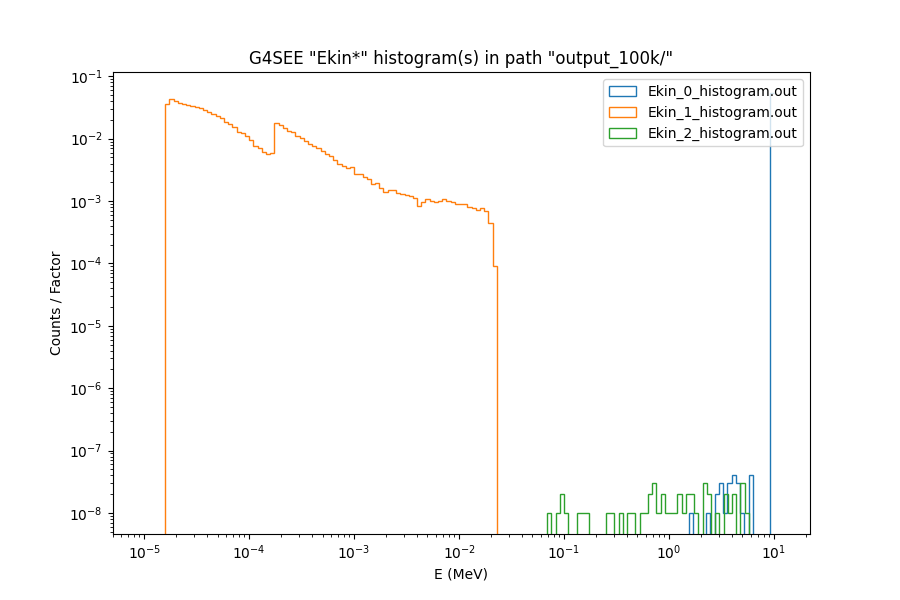
Plotting all particle kinetic energy (Ekin) histograms of a run using g4see.py plot#

